Every story or study needs an introduction to give some context to what we are look at. When I first started writing the introduction about Helicobacter pylori, I realised quite quickly that my first attempts were massively paraphrasing again and again Wikipedia’s summary. Because the Wikipedia article contains most of the basic information I will probably skip some of it and focus more on other interesting points about these bacteria.
However, for those who are absolutely not familiar with our subject, Helicobacter pylori are spiral shaped bacteria found in 30 to 50% of the human population, depending on the region. It gained a lot of attention in the 70’s, when scientist related the H. pylori infection to chronic gastritis and other duodenal cancers. [1]
The pathogenesis of H. pylori has been extensively studied in the past four decades since its correlation to gastric disease. Many studies and reviews [2-3-4] have looked at which proteins are involved in the infections. Fast-forwarding through the history of discoveries leading to the understanding of the molecular mechanisms behind the pathogenicity of H. pylori, we now know that several genes are involved in high strain virulence, gastritis and other duodenal cancers. Going in order of “appearance” during the infection, the most important genes are: babA – a bacterial adhesin that allows sticking to the gut epithelia, cagA – general disruption of the host cell, inhibition of the cell apoptosis and enhancement of cell proliferation, and vacA – a vacuole inducing protein to steal nutrient from the host, as well as various immunity inhibitors and many other genes with local roles. However, even if those proteins have an important role in infection and virulence, some cases of H. pylori-induced gastritis or cancer have been shown to lack some or all of the above-mentioned genes.
Try to be original...
As I finally got to writing my H. pylori adventure to be published on my blog, I found a recent publication from Thorell et al (2017). They looked at 401 H. pylori genomes and showed very nice biogeographical distribution and evolution of different strains in the Americas. Check it out!
My blog article will probably cover some of the content of this publication, but let’s see what the differences and similarities are.
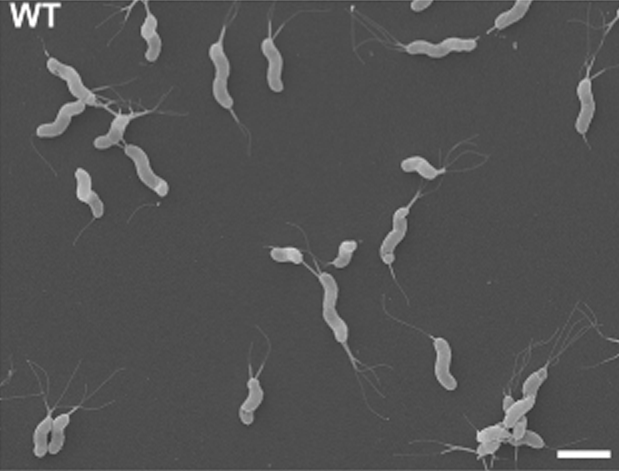
This is one nice H. pylori SEM picture from Sycuro et al, 2013.
Note: It was unexpectedly difficult to find a free of right nice EM picture of H. pylori.
The prevalence of H. pylori in the human population greatly depends on geographical and (probably) socio-economic settings. Database surveys in 2014 showed a variation in the prevalence of the bacteria from it being present in a third of the population in North America and northern Europe to up to half the population in South America and southern and eastern Europe. In certain Asian and African countries as high as 70 to 80 % of the population carried the bacteria [2]. However, the same study indicated that the bacterial prevalence is declining in recent generation in northern countries.
The transmission mode of the bacteria is still unclear, although there is indication of vertical transmission, such as familial and faecal to oral transmissions. Additionally, crowded living conditions, poor sanitation, poor personal hygiene and a poor water supply correlate with higher rates of infection [5]. The settlement of H. pylori appears to be during childhood and the infection of adults still remains unclear. [4]
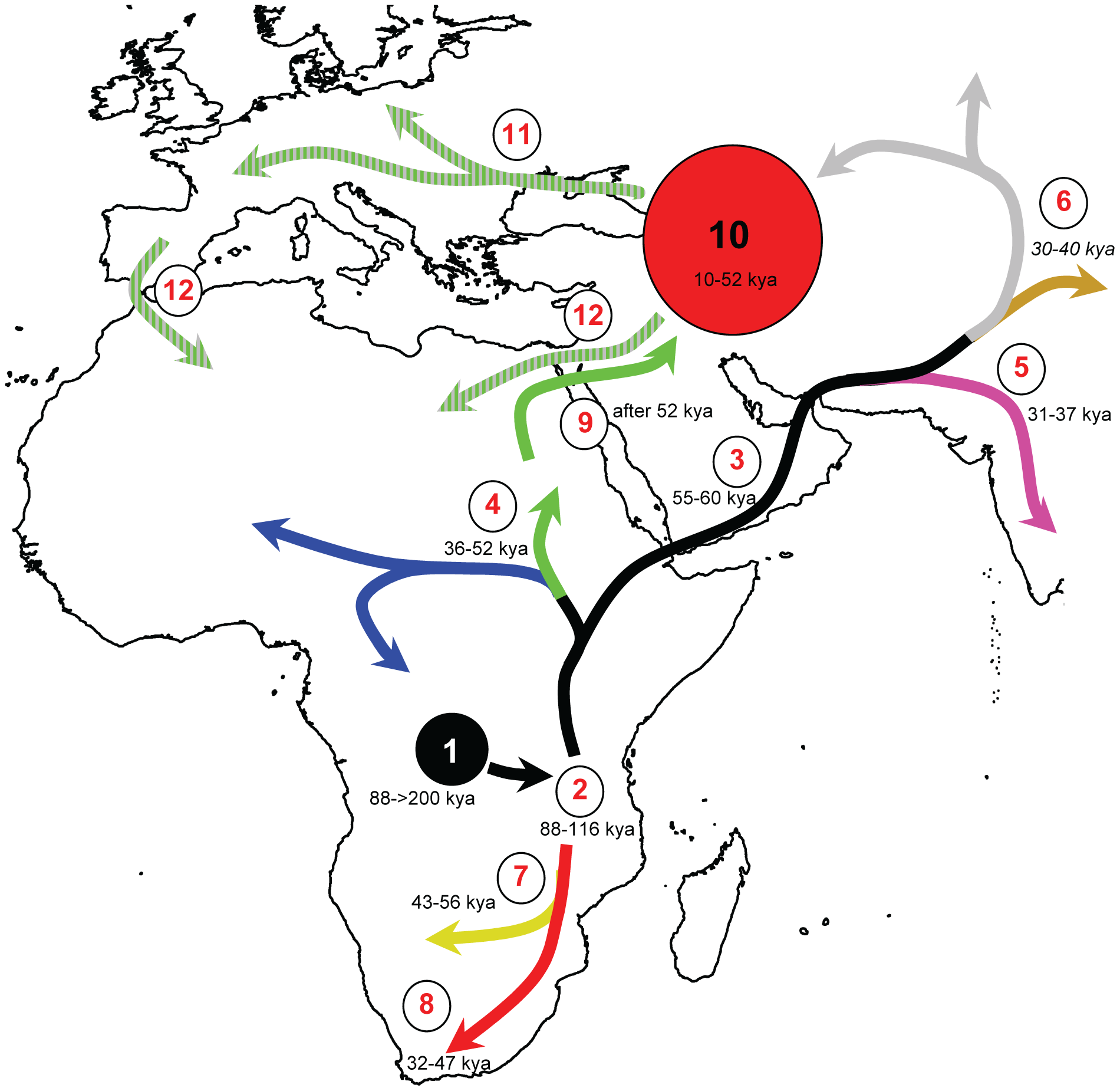
From the intimate association of H. pylori and humans comes one of the most interesting features of this story: the close coevolution of humans and H. pylori. Thought to be acquired by human ancestors more than 100,000 years ago, H. pylori followed human migration and has a strong biogeographical distribution around the world. The initial strain is predicted to have been acquired by a single host jump somewhere in Africa, then diverged into two distinct lineages, one going south and one going north. The northern strain eventually evolved into many other regional ones with human settling in Europe, Asia and further. [6]
Human history and massive population displacement have an impact on the biogeographical distribution of H. pylori. This is noticeable in the H. pylori strain distribution in the Americas. The two mains event of its spreading were the Spanish invasion of South America and the slave trade, which deported millions of Africans to northern America. [7] Nowadays the European and African strains are more abundant in the new world than strains related to the native Amerindian ancestors [8]. Perhaps this geographical pattern will progressively disappear or evolve in a different way, especially when considering our modern society and increasingly easy population movement across the globe. Just thinking of it, I grew up between Burma, China, France and French Guiana, I wonder which pylori I have.
With this succinct introduction, I hope to have set up the two driving questions of this exercise. Using the genomes available on NCBI, I will first have a look at the phylogenomic and geographical distribution of the strains and then look at the gene presence/absence in the different strains. I will certainly also have a look at orthologs and pan-genomes, maybe even look at genome synteny.
Stay tuned, this is only the beginning. Do not hesitate to leave a comment if I missed something important or misinterpreted any data.
References
- Ernst, P. B., and B. D. Gold. (2000). The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu. Rev. Microbiol. 54:615-640. doi:10.1146/annurev.micro.54.1.615
- Johannes G. Kusters, Arnoud H. M. van Vliet and Ernst J. Kuipers (2006) Pathogenesis of Helicobacter pylori Infection. Clin. Microbiol. Rev. vol. 19 no. 3 449-490 doi: 10.1128/CMR.00054-05
- De Falco, M., Lucariello, A., Iaquinto, S., Esposito, V., Guerra, G. and De Luca, A. (2015), Molecular Mechanisms of Helicobacter pylori Pathogenesis. J. Cell. Physiol., 230: 1702–1707. doi:10.1002/jcp.24933
- Backert, S., Neddermann, M., Maubach, G. and Naumann, M. (2016), Pathogenesis of Helicobacter pylori infection. Helicobacter, 21: 19–25. doi:10.1111/hel.12335
- Eusebi, L. H., Zagari, R. M. and Bazzoli, F. (2014), Epidemiology of Helicobacter pylori Infection. Helicobacter, 19: 1–5. doi:10.1111/hel.12165
- Moodley Y, Linz B, Bond RP, Nieuwoudt M, Soodyall H, Schlebusch CM, et al. (2012) Age of the Association between Helicobacter pylori and Man. PLoS Pathog 8(5): e1002693. doi:10.1371/journal.ppat.1002693
- Correa P, Piazuelo MB. Evolutionary History of the Helicobacter pylori Genome: Implications for Gastric Carcinogenesis. Gut and Liver. 2012;6(1):21-28. doi:10.5009/gnl.2012.6.1.21.
- Domínguez-Bello MG, Pérez ME, Bortolini MC, Salzano FM, Pericchi LR, Zambrano-Guzmán O, et al. (2008) Amerindian Helicobacter pylori Strains Go Extinct, as European Strains Expand Their Host Range. PLoS ONE 3(10): e3307. doi:10.1371/journal.pone.0003307
F1. Sycuro, L. K., Rule, C. S., Petersen, T. W., Wyckoff, T. J., Sessler, T., Nagarkar, D. B., Khalid, F., Pincus, Z., Biboy, J., Vollmer, W. and Salama, N. R. (2013), Flow cytometry-based enrichment for cell shape mutants identifies multiple genes that influence Helicobacter pylori morphology. Molecular Microbiology, 90: 869–883. doi:10.1111/mmi.12405