Hello Folks, it’s been a while since I posted here. Plenty of things have happened since my previous Japan posts. The main one being the new postdoc position I started this March in the lab of Buck Samuel at Baylor College of Medicine in Houston, Texas.
Reducing your life to 5 boxes, moving across an Ocean, going through (once more) the paperwork of a new place, resettling in a new flat (I can’t remember how many toilet brushes and cutlery sets I have bought in my life, but I am sure it’s above average), getting used to a new country, a new city, a new lab and getting in a new routine. Hopefully a new one.
Among the good resolutions of this new postdoc is first to adopt a more coherent log of my work, thoughts and ideas than previously. I am currently using the Jupyter Notebook and I am quite satisfied with it, so far. I’ll probably blog about this more specifically later. The second resolution is to try to have a weekly blog post about a paper, forcing me to keep a regular lit review habit and this blog alive at the same time.
The first paper I would like to expand about is the very recently published: “The social network of microorganisms — how auxotrophies shape complex communities” by Karsten Zengler & Livia S. Zaramela from the University of California. (You can find the paper here – Not open)
It’s not very often that I find a paper that includes so many topics I like: Microbiology, Host-microbe interactions, Microbe-microbe interactions, Interaction networks, Correlative analyses and the potential plethora of omics application to solve all the questions going with it.
This paper is a “Perspective opinion” piece in Nature Microbiology Reviews focusing mainly on the need to analyze complex bacterial communities and understand how each member interacts with each other. The paper title includes the sexy buzz words of Social Network, which it is, but does not really go in depth in the full details of a social network and choose to focus instead on the amino acid and vitamin interactions. It’s fair enough, this topic is very broad and has many, many, many sides.

I like this paper because it starts by highlighting the deep complexity of microbial communities, a concept I had in mind for a long time and I think it is clear for a lot of other microbiologists. A microbial population is more than just a simple food web of who eats/transforms which sugar to the benefit of whom. There are many more aspects and potential interactions, such as:
- Amino Acids and Vitamins (discussed in the present paper).
- Detoxification/Inhibition/Inactivation of deleterious molecules. (I don’t like this, maybe you too)
- Production of deleterious molecules. (I don’t want you to grow here)
- Modification of the local environment. (I adjust the pH for myself, maybe you’ll like it)
- Modulation of the host. (I just tricked the host cell to release calcium, maybe you’ll like it too)
- All the other I can’t think of right now.
- And finally all the other we don’t know about yet.
When we look at as such an interaction mess, how do we understand how it works? Where do we start?
Well, the first thing to do was indeed to look at the simple food web. To see what makes the community stable, we need first to understand what makes each of its member happy. It is the logical way, first understand the type of carbon and energy sources needed by the bacteria, and then look if anyone else would profit from it. On that point, the paper makes a nice explanation of the traditional way to approach to explore community networks by looking at the fate of main nutrients, such as carbon source and electron sources.
But then what about the rest? The paper states that 98% of sequenced bacterial genomes are missing pathways or key genes for at least one type of amino acid. And up until now, we often don’t know where the missing amino acid for one specific community member comes from. Does it come from decaying cells in the environment? An overflow of another member? Active efflux from another member? The host? Something we don’t know yet?
And it doesn’t stop there… If two members of the community have redundant main metabolic pathways, do they have the same amino acid biosynthesis repertoire? And if not, where do they get it and which member gets the most out of it? How does it vary in the long run? The paper includes several examples of synthetic communities and simple systems and challenges the field for the study of more complex systems which include a high diversity of bacteria and/or a host. It also underlines the competition for those amino acids within the community, how do different member needing the same amino acids acquire it and how do they keep acquiring it? It is also a discussed point on how a settled microbiota is actually preventing any new member (pathogen) to settle in: by keeping the good molecule for themselves.
That’s the exciting points of the paper. Plenty of question, plenty of work for the present and future microbiome researcher. The discussion comes naturally to use the plethora of novel techniques to finely dissect microbial communities. And in an ideal world you would throw money at a multi-omics approach where you can do metagenomes/transcriptomes on longitudinal experiments and explore the variation of microbial members across time, but not only, you could add to this metabolomics, and particularly the field of imaging metabolomics and fine tune the spatial distribution of metabolites within a community across your experiment and link it with the previous omic data.
Another point they discuss and I would like to underline too is the transporter rabbit hole. This is something we had a lot of trouble to deal with in my previous lab, how to properly annotate transporters. And how to interpret it correctly. The short answer is with great difficulty, because very little is know about the specificity, ubiquity and mechanism of bacterial transporters and all of this is linked to poor annotation/interpretations. I think the best example of the paper is this one:
Bacteria are automates, with actions defined/limited by their genome. We are slowly learning all the moves they can make, it is now (or very soon) time to figure out how the game is played.
This paper is good and bad news. The good news is that there is work for everyone here and coming to the microbiome world, so many questions which lead to so many others. The bad one, which is not that bad but still, is that with a paper like this out, figuring out the complexity of microbiome is not anything new. It is still exiting, absolutely necessary to be done but this part of the concept has been laid down, the water is murky. To keep the research exciting and groundbreaking we need to find the next clear pool to jump in!
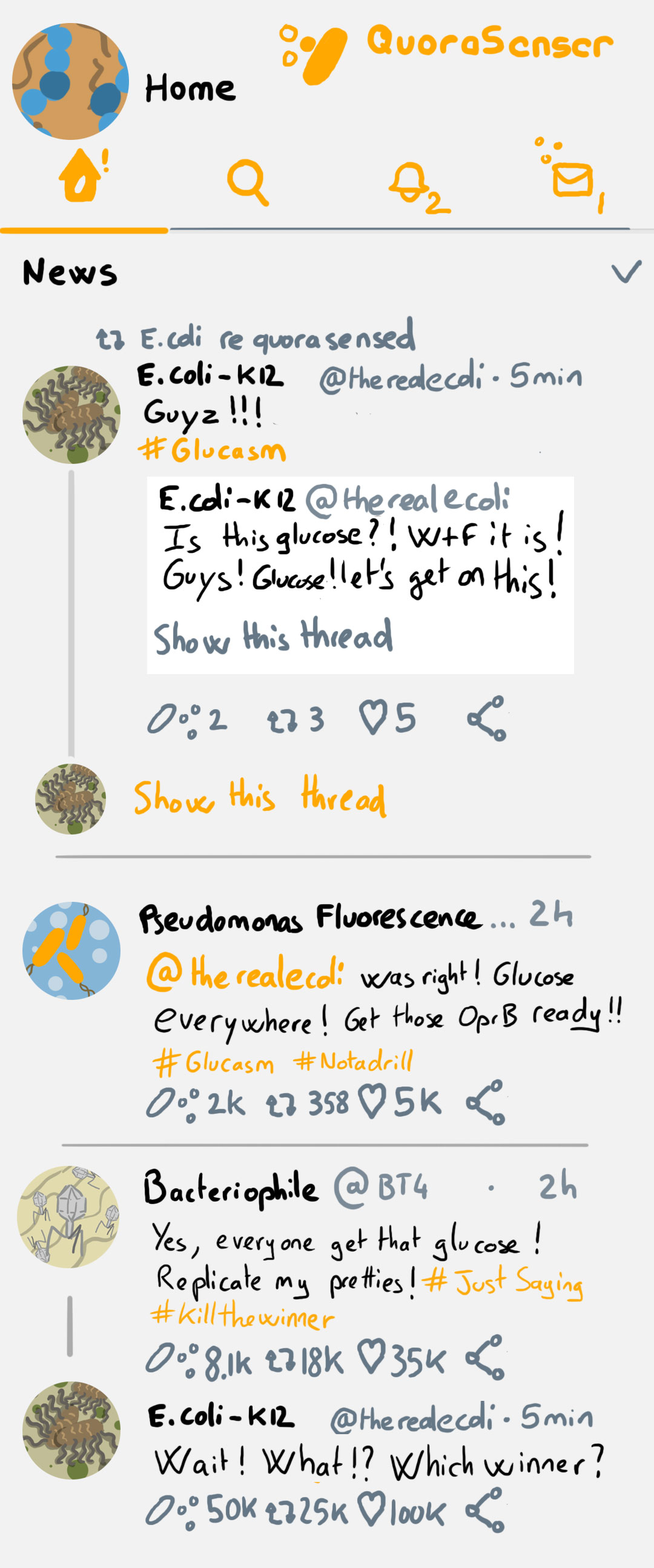
While we find it, enjoy a mock Bacterial Social Network on the left 🙂
Also getting into the habit of a regulat lit routine again ^^ Awesome illustrations!