I am sitting on this blog for too long. I think it is time I try to publish something useful on it. So to do so I am going to present here in a series of short articles how I handle metagenome analyses. I am not pretending to have the one and only solution, I just think it will be a nice to have it nicely documented somewhere (in case I ever forget how to do it) and if it can help someone in the same time why not.
This first article is going to describe what the early step of preparing raw read files before the assembly phase. I will present my process using publicly available metagenome from the Human Metagenome Project. I have selected the first metagenome available from the tongue dorsum sample category.
Let’s have a look a the bacterial community of the tongue of the female participant SRS147120.
This is a 100 bp paired end read library sequenced with the Illumina technology.

After downloading the raw reads from the HMP ftp, I extracted the reads from the bz2 format. They will be recompress on the next step to a more useful .gz format.
bzip2 -d SRS147120.tar.bz2
tar xvf SRS147120.tar
We can first have a look at the library quality, to do so we can use the FastQC software which output us with a summary looking like this:
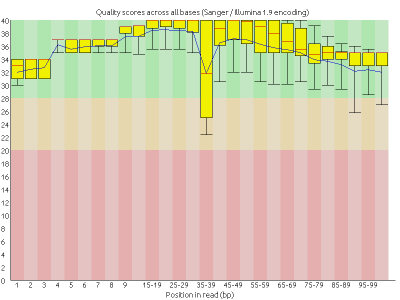
This indicates that overall the quality of the reads is good. On the middle of the sequence we can observe a drop in quality which is apparently due to a insertion of N. A thing to do is to clean the reads of bad quality reads and other sequencing artifacts (PhiX sequences, primers, etc…) using bbduk from Brian Bushnell’s tool box: BBmap.
bbduk.sh in=SRS147120/SRS147120.denovo_duplicates_marked.trimmed.1.fastq in2=SRS147120/SRS147120.denovo_duplicates_marked.trimmed.2.fastq ref=/tool/bbmap/resources/adapters.fa,/tool/bbmap/resources/phix_adapters.fa.gz,/tool/bbmap/resources/truseq.fa.gz trimq=2 qtrim=rl out=reads_q2.fq.gz
We are inputing the forward and reverse reads with in and in2, with the ref option we ask the script to check a list of known contaminant of the illumina pipeline, trimq and qtrim options are going to trim read regions with a bad quality. Finally we are going to output the screened reads in an interleaved and gz compressed format. This is what the output looks like.
BBDuk version 35.59
Initial:
Memory: max=7448m, free=7292m, used=156m
Added 2970 kmers; time: 0.088 seconds.
Memory: max=7448m, free=6787m, used=661m
Input is being processed as paired
Started output streams: 0.025 seconds.
Processing time: 157.832 seconds.
Input: 92263798 reads 9226379800 bases.
Contaminants: 47440 reads (0.05%) 4744000 bases (0.05%)
Low quality discards: 0 reads (0.00%) 0 bases (0.00%)
Result: 92216358 reads (99.95%) 9221635800 bases (99.95%)
Time: 157.952 seconds.
Reads Processed: 92263k 584.12k reads/sec
Bases Processed: 9226m 58.41m bases/sec
We had apparently little bad apple in the assembly (0,005% the early quality check of the HMP people is efficient). What we can do now is to use a RNA subunit screening tool PhyloFlash. This tool is currently developed by one of my college: Harald Gruber-Vodicka. It allows to have a idea of what organism to expect from a metagenome/metatranscriptomic library. This is very useful to see if you have potential contamination in your library. (For example if you have butterfly sequences in a deep sea sample, it might happen). We run the following command:
phyloFlash.pl -lib tongue -read1 reads_q2.fq.gz -CPUs 5
Here the lib option call the name you want to prefix the outputted files, read1 your input file (can be read compressed), and CPUs call the number of processors you want to use. (Note if you ask too many processors, the process might crash during one of the steps because it distribute a fixed memory amount between the available cpus).
Having a look at the outputted file tongue.phyloFlash will tell you what taxa you can expect in your library. In our case, the metagenome seems to be host of a large diversity of different bacterial taxa. The output file is placed in the following collapsed section.
Section
phyloFlash v2.0 – high throughput phylogenetic screening using SSU rRNA gene(s) abundance(s)
Library name: tongue
—
Forward read file reads_q2.fq.gz
Reverse read file <NONE>
Current working directory /opt/extern/bremen/molecol/aassie/000_blog
—
Minimum mapping identity: 70%
—
Input PE-reads: 92216358
Mapped SSU read pairs: 97729
Mapping ratio: 0.105977943739656%
Detected median insert size: 202
Used insert size: SE mode!
Insert size standard deviation: 412
—
Runtime: 202.52 minutes
CPUs used: 5
—
Read mapping based higher taxa (NTUs) detection
NTUs observed once: 70
NTUs observed twice: 31
NTUs observed three or more times: 101
NTU Chao1 richness estimate: 180.032258064516
List of NTUs in order of abundance:
NTU reads
Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella 2453
Bacteria;Proteobacteria;Betaproteobacteria;Burkholderiales;Comamonadaceae;Caenimonas 1635
Bacteria;Proteobacteria;Betaproteobacteria;Neisseriales;Neisseriaceae;Neisseria 1199
Bacteria;Actinobacteria;Actinobacteria;Actinomycetales;Actinomycetaceae;Actinomyces 1086
Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus 905
Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella 7 855
Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Porphyromonadaceae;Porphyromonas 799
Bacteria;Proteobacteria;Gammaproteobacteria;Legionellales;Legionellaceae;Legionella 651
Bacteria;Firmicutes;Negativicutes;Selenomonadales;Veillonellaceae;Veillonella 621
Bacteria;Fusobacteria;Fusobacteriia;Fusobacteriales;Leptotrichiaceae;Leptotrichia 466
Bacteria;Proteobacteria;Gammaproteobacteria;Pasteurellales;Pasteurellaceae;Haemophilus 436
Bacteria;Actinobacteria;Actinobacteria;Micrococcales;Micrococcaceae;Rothia 389
Bacteria;Firmicutes;Clostridia;Clostridiales;Lachnospiraceae;Stomatobaculum 384
Bacteria;Firmicutes;Erysipelotrichia;Erysipelotrichales;Erysipelotrichaceae;Solobacterium 334
Bacteria;Fusobacteria;Fusobacteriia;Fusobacteriales;Fusobacteriaceae;Fusobacterium 251
Bacteria;Proteobacteria;Epsilonproteobacteria;Campylobacterales;Campylobacteraceae;Campylobacter 241
Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Alloprevotella 228
Bacteria;Firmicutes;Clostridia;Clostridiales;Ruminococcaceae;Ruminococcaceae UCG-014 210
Bacteria;Firmicutes;Clostridia;Clostridiales;Lachnospiraceae;Oribacterium 204
Bacteria;Firmicutes;Erysipelotrichia;Erysipelotrichales;Erysipelotrichaceae;Erysipelotrichaceae UCG-007 200
Bacteria;Actinobacteria;Actinobacteria;Streptomycetales;Streptomycetaceae;Streptomyces 123
Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacteriales;Enterobacteriaceae;Escherichia-Shigella 118
Bacteria;Firmicutes;Clostridia;Clostridiales;Family XIII;Mogibacterium 106
Bacteria;Tenericutes;Mollicutes;Mollicutes RF9;Firmicutes oral clone FM046 95
Bacteria;Firmicutes;Clostridia;Clostridiales;Lachnospiraceae;Lachnoanaerobaculum 92
Bacteria;Firmicutes;Clostridia;Clostridiales;Peptostreptococcaceae;Peptostreptococcus 91
Bacteria;Firmicutes;Clostridia;Clostridiales;Lachnospiraceae;[Eubacterium] oxidoreducens group 87
Bacteria;Saccharibacteria;uncultured bacterium 82
Bacteria;Firmicutes;Bacilli;Bacillales;Family XI;Gemella 81
Bacteria;Firmicutes;Negativicutes;Selenomonadales;Veillonellaceae;Megasphaera 80
Bacteria;Firmicutes;Bacilli;Lactobacillales;Lactobacillaceae;Lactobacillus 77
Bacteria;Firmicutes;Clostridia;Clostridiales;Lachnospiraceae;Butyrivibrio 2 75
Bacteria;Saccharibacteria;Unknown Class;Unknown Order;Unknown Family;Candidatus Saccharimonas 75
Bacteria;Firmicutes;Bacilli;Lactobacillales;Carnobacteriaceae;Granulicatella 74
Bacteria;Actinobacteria;Actinobacteria;Corynebacteriales;Mycobacteriaceae;Mycobacterium 67
Bacteria;Firmicutes;Clostridia;Clostridiales;Family XIII;[Eubacterium] nodatum group 65
Bacteria;Actinobacteria;Coriobacteriia;Coriobacteriales;Coriobacteriaceae;Atopobium 59
Bacteria;Saccharibacteria;TM7 phylum sp. oral clone DR034 58
Bacteria;Proteobacteria;Betaproteobacteria;Neisseriales;Neisseriaceae;Alysiella 57
Bacteria;Firmicutes;Negativicutes;Selenomonadales;Veillonellaceae;Selenomonas 3 54
Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella 6 53
Bacteria;Proteobacteria;Betaproteobacteria;Neisseriales;Neisseriaceae;uncultured 51
Bacteria;Firmicutes;Clostridia;Clostridiales;Peptostreptococcaceae;Filifactor 47
Bacteria;Firmicutes;Clostridia;Clostridiales;Lachnospiraceae;uncultured 43
Bacteria;Bacteroidetes;Flavobacteriia;Flavobacteriales;Flavobacteriaceae;Capnocytophaga 43
Bacteria;Firmicutes;Clostridia;Clostridiales;Lachnospiraceae;Catonella 41
Bacteria;Firmicutes;Clostridia;Clostridiales;Family XI;Parvimonas 38
Bacteria;Cyanobacteria;Chloroplast;Aegilops tauschii 35
Bacteria;Firmicutes;Bacilli;Bacillales;Bacillaceae;Bacillus 34
Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacteriales;Enterobacteriaceae;Salmonella 33
Bacteria;Bacteroidetes;Cytophagia;Cytophagales;Cyclobacteriaceae;Rhodonellum 31
Bacteria;Saccharibacteria;Candidatus Saccharibacteria oral taxon TM7x 25
Bacteria;Saccharibacteria;TM7 phylum sp. oral clone FR058 23
Bacteria;Firmicutes;Clostridia;Clostridiales;Peptostreptococcaceae;Peptoclostridium 21
Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacteriales;Enterobacteriaceae;Enterobacter 19
Bacteria;Firmicutes;Bacilli;Bacillales;Staphylococcaceae;Staphylococcus 17
Bacteria;Bacteroidetes;Flavobacteriia;Flavobacteriales;Flavobacteriaceae;Bergeyella 16
Bacteria;FirmicutesBacteria;Firmicutes;Clostridia;Clostridiales;Ruminococcaceae;[Eubacterium] coprostanoligenes group 10
Bacteria;Synergistetes;Synergistia;Synergistales;Synergistaceae;Fretibacterium 9
Bacteria;Proteobacteria;Betaproteobacteria;Neisseriales;Neisseriaceae;Kingella 9
Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotellaceae UCG-003 9
Bacteria;Firmicutes;Bacilli;Lactobacillales;Aerococcaceae;Abiotrophia 8
Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella 9 8
Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella 1 8
Bacteria;Actinobacteria;Actinobacteria;Corynebacteriales;Corynebacteriaceae;Corynebacterium 8
Bacteria;Firmicutes;Bacilli;Bacillales;Bacillaceae;Salirhabdus 7
Bacteria;Proteobacteria;Betaproteobacteria;Neisseriales;Neisseriaceae;Eikenella 7
Bacteria;Proteobacteria;Betaproteobacteria;Burkholderiales;Comamonadaceae;Comamonas 7
Bacteria;Firmicutes;Bacilli;Lactobacillales;Enterococcaceae;Vagococcus 6
Bacteria;Actinobacteria;Acidimicrobiia;Acidimicrobiales;Acidimicrobiaceae;Illumatobacter 6
Bacteria;Firmicutes;Bacilli;Lactobacillales;P5D1-392;uncultured bacterium 6
Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacteriales;Enterobacteriaceae;Serratia 6
Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Porphyromonadaceae;Tannerella 6
Bacteria;Proteobacteria;Gammaproteobacteria;Cardiobacteriales;Cardiobacteriaceae;Cardiobacterium 5
Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Bacteroidaceae;Bacteroides 5
Bacteria;Cyanobacteria;Chloroplast;Coccomyxa sp. LA000219 5
Bacteria;Firmicutes;Negativicutes;Selenomonadales;Veillonellaceae;Dialister 5
Bacteria;Proteobacteria;Gammaproteobacteria;Pasteurellales;Pasteurellaceae;Basfia 5
Bacteria;Proteobacteria;Betaproteobacteria;Neisseriales;Neisseriaceae;Vogesella 5
Bacteria;Proteobacteria;Betaproteobacteria;Burkholderiales;Burkholderiaceae;Lautropia 5
Bacteria;Proteobacteria;Betaproteobacteria;Neisseriales;Neisseriaceae;Snodgrassella 4
Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Bacteroidales S24-7 group;uncultured bacterium 4
Bacteria;Fusobacteria;Fusobacteriia;Fusobacteriales;Leptotrichiaceae;Sebaldella 4
Bacteria;Proteobacteria;Gammaproteobacteria;Pasteurellales;Pasteurellaceae;Actinobacillus 4
Bacteria;Proteobacteria;Gammaproteobacteria;Vibrionales;Vibrionaceae;Vibrio 4
Bacteria;Saccharibacteria;candidate division TM7 bacterium JGI 0001002-L20 4
Bacteria;Fusobacteria;Fusobacteriia;Fusobacteriales;CFT112H7;uncultured bacterium 3
Bacteria;Firmicutes;Bacilli;Lactobacillales;Enterococcaceae;Enterococcus 3
Bacteria;Proteobacteria;Gammaproteobacteria;Pasteurellales;Pasteurellaceae;Aggregatibacter 3
Bacteria;Proteobacteria;Alphaproteobacteria;Sphingomonadales;Erythrobacteraceae;Altererythrobacter 3
Bacteria;Proteobacteria;Gammaproteobacteria;Vibrionales;Vibrionaceae;Salinivibrio 3
Bacteria;Proteobacteria;Betaproteobacteria;Neisseriales;Neisseriaceae;Vitreoscilla 3
Bacteria;Proteobacteria;Gammaproteobacteria;Pasteurellales;Pasteurellaceae;Gallibacterium 3
Bacteria;Actinobacteria;Actinobacteria;Micrococcales;Intrasporangiaceae;Tetrasphaera 3
Bacteria;Firmicutes;Bacilli;Lactobacillales;P5D1-392;unidentified 3
;Bacilli;Bacillales;Planococcaceae;Planomicrobium 16
Bacteria;Firmicutes;Bacilli;Bacillales;Paenibacillaceae;Cohnella 13
Bacteria;Firmicutes;Clostridia;Clostridiales;Lachnospiraceae;Lachnospiraceae UCG-008 12
Bacteria;Proteobacteria;Gammaproteobacteria;Pseudomonadales;Pseudomonadaceae;Pseudomonas 11
Bacteria;Firmicutes;Negativicutes;Selenomonadales;Veillonellaceae;Selenomonas 11
Bacteria;Fusobacteria;Fusobacteriia;Fusobacteriales;ASCC02;Granulicatella sp. oral clone ASC02 10
Bacteria;Firmicutes;Clostridia;Clostridiales;Ruminococcaceae;[Eubacterium] coprostanoligenes group 10
—
SSU assembly based taxa:
OTU coverage dbHit taxonomy %id alnlen evalue
tongue.PFspades_1 163.282 ACQV01000002.9075.10620 Bacteria;Proteobacteria;Betaproteobacteria;Neisseriales;Neisseriaceae;Neisseria;Neisseria flavescens SK114 99.7 1523 -1 99.5 1536 1521 2 3 1518
tongue.PFspades_16 58.4367 HQ777184.1.1470 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;uncultured organism 100.0 1176 -1 99.9 1176 1176 0 0 1176
tongue.PFspades_15 49.3781 JQ449239.1.1395 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Porphyromonadaceae;Porphyromonas;uncultured bacterium 99.8 959 -1 99.7 964 958 1 1 957
tongue.PFspades_2 33.0807 AJTC01000001.52.1592 Bacteria;Proteobacteria;Gammaproteobacteria;Pasteurellales;Pasteurellaceae;Haemophilus;Haemophilus parainfluenzae HK2019 99.6 1520 -1 99.5 1536 1520 0 6 1514
tongue.PFspades_5 23.7396 GQ338720.1.1529 Bacteria;Firmicutes;Negativicutes;Selenomonadales;Veillonellaceae;Veillonella;uncultured bacterium 98.3 1529 -1 98.2 1564 1527 2 24 1503
tongue.PFspades_3 20.5142 HK240338.10.1530 Bacteria;Actinobacteria;Actinobacteria;Actinomycetales;Actinomycetaceae;Actinomyces;unidentified 98.9 1521 -1 98.8 1538 1521 0 16 1505
tongue.PFspades_10 17.6272 ACJY01000002.122.1615 Bacteria;Fusobacteria;Fusobacteriia;Fusobacteriales;Fusobacteriaceae;Fusobacterium;Fusobacterium periodonticum ATCC 33693 99.8 1486 -1 99.7 1503 1486 0 3 1483
tongue.PFspades_18 10.3672 JQ461559.1.1411 Bacteria;Firmicutes;Bacilli;Lactobacillales;Carnobacteriaceae;Granulicatella;uncultured bacterium 100.0 957 -1 99.9 961 957 0 0 957
tongue.PFspades_7 9.84772 HK241843.9.1513 Bacteria;Firmicutes;Erysipelotrichia;Erysipelotrichales;Erysipelotrichaceae;Solobacterium;unidentified 99.6 1505 -1 99.5 1522 1505 0 6 1499
tongue.PFspades_4 9.64834 ACSB01000014.3447.4963 Bacteria;Actinobacteria;Actinobacteria;Micrococcales;Micrococcaceae;Rothia;Rothia mucilaginosa M508 99.8 1509 -1 99.7 1526 1509 0 3 1506
tongue.PFspades_14 8.7575 KC169767.1.1488 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Alloprevotella;uncultured Bacteroidetes bacterium 99.8 959 -1 99.7 963 959 0 2 957
tongue.PFspades_8 8.19441 AF385520.1.1496 Bacteria;Saccharibacteria;TM7 phylum sp. oral clone DR034 99.5 1490 -1 99.3 1525 1490 0 8
1482
tongue.PFspades_11 6.41068 AENQ01000044.3766.5256 Bacteria;Proteobacteria;Epsilonproteobacteria;Campylobacterales;Campylobacteraceae;Campylobacter;Campylobacter concisus UNSWCD 99.9 1491 -1 99.8 1508 1491 0 1 1490
tongue.PFspades_9 4.48011 AWVR01000095.17.1514 Bacteria;Fusobacteria;Fusobacteriia;Fusobacteriales;Leptotrichiaceae;Leptotrichia;Leptotrichia sp. oral taxon 215 str. W9775 99.4 1490 -1 99.3 1507 1490 0 9 1481
tongue.PFspades_6 4.00532 AF287763.1.1537 Bacteria;Firmicutes;Clostridia;Clostridiales;Peptostreptococcaceae;Peptostreptococcus;Peptostreptococcus sp. oral clone CK035 99.7 1523 -1 99.6 1541 1523 0 4 1519
tongue.PFspades_13 3.01362 AF385510.1.1522 Bacteria;Firmicutes;Clostridia;Clostridiales;Lachnospiraceae;Stomatobaculum;Eubacterium sp. oral clone DO016 95.5 1514 -1 95.8 1530 1506 8 60 1446
tongue.PFspades_12 1.73785 HE681253.1.1518 Bacteria;Firmicutes;Clostridia;Clostridiales;Ruminococcaceae;Ruminococcaceae UCG-014;uncultured bacterium 99.5 1509 -1 99.3 1526 1509 0 8 1501
tongue.PFspades_17 1.65886 JQ465246.1.1375 Bacteria;Firmicutes;Clostridia;Clostridiales;Family XIII;Mogibacterium;uncultured bacterium 99.9 972 -1 99.8 978 972 0 1 971
—
SSU reconstruction based taxa:
OTU ratio dbHit taxonomy %id alnlen evalue
tongue.PFemirge_7 0.196360 JQ449243.1.1398 Bacteria;Proteobacteria;Betaproteobacteria;Neisseriales;Neisseriaceae;Neisseria;uncultured bacterium 97.4 1401 -1 97.4 1401 1398 3 34 1364
tongue.PFemirge_31 0.058397 JQ460126.1.1415 Bacteria;Firmicutes;Negativicutes;Selenomonadales;Veillonellaceae;Veillonella;uncultured bacterium 99.9 1314 -1 99.9 1314 1314 0 1 1313
tongue.PFemirge_169 0.057802 FM995684.1.1473 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Porphyromonadaceae;Porphyromonas;uncultured bacterium 99.7 1473 -1 99.5 1510 1473 0 5 1468
tongue.PFemirge_10 0.046183 JQ449266.1.1397 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella 7;uncultured bacterium 99.8 1390 -1 99.6 1417 1390 0 3 1387
tongue.PFemirge_350 0.042717 EF511181.1.1499 Bacteria;Proteobacteria;Gammaproteobacteria;Pasteurellales;Pasteurellaceae;Haemophilus;uncultured bacterium 99.2 1481 -1 99.2 1481 1481 0 12 1469
tongue.PFemirge_572 0.030953 JQ449672.1.1409 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;uncultured bacterium 99.3 1409 -1 99.3 1409 1409 0 10 1399
tongue.PFemirge_88 0.022259 AJTC01000001.52.1592 Bacteria;Proteobacteria;Gammaproteobacteria;Pasteurellales;Pasteurellaceae;Haemophilus;Haemophilus parainfluenzae HK2019 98.8 1515 -1 99.4 1515 1504 11 7 1497
tongue.PFemirge_93 0.021629 JQ451880.1.1378 Bacteria;Fusobacteria;Fusobacteriia;Fusobacteriales;Fusobacteriaceae;Fusobacterium;uncultured bacterium 96.7 1379 -1 96.8 1377 1375 4 41 1334
tongue.PFemirge_111 0.021404 AF366269.1.1534 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella 7;uncultured Prevotella sp. 99.5 1517 -1 99.5 1517 1517 0 7 1510
tongue.PFemirge_192 0.021016 JQ460482.1.1399 Bacteria;Proteobacteria;Betaproteobacteria;Neisseriales;Neisseriaceae;Neisseria;uncultured bacterium 99.8 1399 -1 99.8 1399 1399 0 3 1396
tongue.PFemirge_9782 0.020839 AEVD01000030.72.1605 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;Streptococcus infantis ATCC 700779 80.4 1554 -1 83.8 1538 1487 67 238 1249
tongue.PFemirge_207 0.020479 KF101920.1.1362 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella;uncultured bacterium 99.7 1362 -1 99.7 1362 1362 0 4 1358
tongue.PFemirge_25 0.019135 AF385504.1.1539 Bacteria;Actinobacteria;Actinobacteria;Actinomycetales;Actinomycetaceae;Actinomyces;Actinomyces sp. oral clone CT047 99.0 1526 -1 99.0 1526 1525 1 14 1511
tongue.PFemirge_15 0.016818 HK241642.1.1511 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella;unidentified 99.7 1511 -1 99.7 1511 1511 0 4 1507
tongue.PFemirge_373 0.015839 AB547695.1.1489 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella;Prevotella nanceiensis 99.4 1398 -1 99.4 1398 1398 0 8 1390
tongue.PFemirge_197 0.015737 JQ449272.1.1394 Bacteria;Proteobacteria;Gammaproteobacteria;Pasteurellales;Pasteurellaceae;Haemophilus;uncultured bacterium 99.8 1394 -1 99.8 1394 1394 0 3 1391
tongue.PFemirge_9708 0.015438 ACQV01000002.9075.10620 Bacteria;Proteobacteria;Betaproteobacteria;Neisseriales;Neisseriaceae;Neisseria;Neisseria flavescens SK114 95.8 1511 -1 95.9 1509 1509 2 61 1448
tongue.PFemirge_610 0.014278 HK241843.9.1513 Bacteria;Firmicutes;Erysipelotrichia;Erysipelotrichales;Erysipelotrichaceae;Solobacterium;unidentified 99.6 1505 -1 99.6 1505 1505 0 6 1499
tongue.PFemirge_719 0.012764 HQ778326.1.1469 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;uncultured organism 96.2 1465 -1 96.5 1461 1457 8 47 1410
tongue.PFemirge_180 0.012558 KF098306.1.1368 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;uncultured bacterium 99.0 1368 -1 99.0 1368 1368 0 13 1355
tongue.PFemirge_1166 0.012404 AF385520.1.1496 Bacteria;Saccharibacteria;TM7 phylum sp. oral clone DR034 99.5 1490 -1 99.5 1490 1490 0
7 1483
tongue.PFemirge_9825 0.011649 AEVD01000030.72.1605 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;Streptococcus infantis ATCC 700779 87.7 1551 -1 91.0 1538 1490 61 130 1360
tongue.PFemirge_126 0.011402 FJ470572.1.1411 Bacteria;Firmicutes;Clostridia;Clostridiales;Lachnospiraceae;[Eubacterium] oxidoreducens group;uncultured bacterium 96.5 1362 -1 96.8 1358 1354 8 40 1314
tongue.PFemirge_675 0.011261 AEVD01000030.72.1605 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;Streptococcus infantis ATCC 700779 86.8 1553 -1 90.2 1538 1488 65 140 1348
tongue.PFemirge_1236 0.011060 FM872752.1.1502 Bacteria;Firmicutes;Bacilli;Lactobacillales;Carnobacteriaceae;Granulicatella;uncultured bacterium 99.9 1458 -1 99.8 1461 1458 0 1 1457
tongue.PFemirge_285 0.011031 GQ338720.1.1529 Bacteria;Firmicutes;Negativicutes;Selenomonadales;Veillonellaceae;Veillonella;uncultured bacterium 98.4 1529 -1 98.4 1529 1529 0 25 1504
tongue.PFemirge_764 0.010882 ACSB01000014.3447.4963 Bacteria;Actinobacteria;Actinobacteria;Micrococcales;Micrococcaceae;Rothia;Rothia mucilaginosa M508 99.4 1451 -1 99.7 1451 1444 7 2 1442
tongue.PFemirge_585 0.009756 AENQ01000044.3766.5256 Bacteria;Proteobacteria;Epsilonproteobacteria;Campylobacterales;Campylobacteraceae;Campylobacter;Campylobacter concisus UNSWCD 99.9 1491 -1 99.9 1491 1491 0 1 1490
tongue.PFemirge_404 0.007731 KC169767.1.1488 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Alloprevotella;uncultured Bacteroidetes bacterium
99.2 1487 -1 99.2 1487 1487 0 12 1475
tongue.PFemirge_100 0.006757 EU531780.1.1546 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;uncultured bacterium 99.2 1528 -1 99.2 1528 1528 0 12 1516
tongue.PFemirge_101 0.006259 FJ558479.1.1379 Bacteria;Actinobacteria;Actinobacteria;Micrococcales;Micrococcaceae;Rothia;uncultured bacterium 96.6 1388 -1 97.6 1379 1370 18 29 1341
tongue.PFemirge_303 0.005992 AY923125.1.1558 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;Streptococcus sp. oral clone ASCB12 98.0 1548 -1 98.1 1544 1540 8 23 1517
tongue.PFemirge_259 0.005546 AY349396.1.1524 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella 6;Prevotella sp. oral clone GI032 99.3 1509 -1 99.2 1509 1509 0 11 1498
tongue.PFemirge_281 0.005305 JN379060.1.1513 Bacteria;Firmicutes;Bacilli;Bacillales;Family XI;Gemella;uncultured bacterium 97.6 1512 -1 97.5 1512 1512 0 37 1475
tongue.PFemirge_613 0.004982 AWVR01000095.17.1514 Bacteria;Fusobacteria;Fusobacteriia;Fusobacteriales;Leptotrichiaceae;Leptotrichia;Leptotrichia sp. oral taxon 215 str. W9775 99.2 1415 -1 99.2 1415 1413 2 9 1404
tongue.PFemirge_488 0.004680 AB637220.1.1505 Bacteria;Firmicutes;Bacilli;Bacillales;Planococcaceae;Planomicrobium;uncultured bacterium 92.0 1507 -1 92.0 1505 1503 4 117 1386
tongue.PFemirge_718 0.004675 FJ470510.1.1487 Bacteria;Bacteroidetes;Flavobacteriia;Flavobacteriales;Flavobacteriaceae;Capnocytophaga;uncultured bacterium 88.9 1492 -1 89.2 1487 1482 10 155 1327
tongue.PFemirge_1 0.004483 HQ778352.1.1429 Bacteria;Fusobacteria;Fusobacteriia;Fusobacteriales;Fusobacteriaceae;Fusobacterium;uncultured organism 98.3 1383 -1 98.2 1383 1382 1 23 1359
tongue.PFemirge_138 0.004463 JQ471598.1.1397 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella 7;uncultured bacterium 99.3 1395 -1 99.2 1395 1395 0 10 1385
tongue.PFemirge_4 0.004402 JQ460226.1.1424 Bacteria;Firmicutes;Negativicutes;Selenomonadales;Veillonellaceae;Veillonella;uncultured bacterium 99.4 1424 -1 99.4 1424 1423 1 7 1416
tongue.PFemirge_390 0.004301 HK241664.1.1506 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Alloprevotella;unidentified 97.8 1505 -1 97.7 1505 1505 0 33 1472
tongue.PFemirge_331 0.004169 JQ449434.1.1369 Bacteria;Firmicutes;Clostridia;Clostridiales;Peptostreptococcaceae;Peptostreptococcus;uncultured bacterium 99.5 1368 -1 99.3 1368 1367 1 6 1361
tongue.PFemirge_190 0.004099 AF385510.1.1522 Bacteria;Firmicutes;Clostridia;Clostridiales;Lachnospiraceae;Stomatobaculum;Eubacterium sp. oral clone DO016 98.5 1510 -1 98.5 1510 1509 1 22 1487
tongue.PFemirge_653 0.004050 AM420032.1.1494 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella 7;uncultured Prevotella sp. 99.0 1493 -1 98.9 1492 1491 2 13 1478
tongue.PFemirge_407 0.004045 JQ460279.1.1390 Bacteria;Actinobacteria;Actinobacteria;Micrococcales;Micrococcaceae;Rothia;uncultured bacterium 74.5 1337 -1 79.6 1307 1265 72 269 996
tongue.PFemirge_747 0.003609 GU361882.1.1515 Bacteria;Firmicutes;Bacilli;Bacillales;Family XI;Gemella;uncultured bacterium 98.2 1515 -1 98.2 1515 1515 0 28 1487
tongue.PFemirge_41 0.003531 HQ771317.1.1450 Bacteria;Proteobacteria;Betaproteobacteria;Neisseriales;Neisseriaceae;Neisseria;uncultured organism 92.2 1315 -1 93.7 1311 1296 19 84 1212
tongue.PFemirge_2433 0.003313 GQ091776.1.1369 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;uncultured bacterium 97.7 1367 -1 97.7 1366 1365 2 29 1336
tongue.PFemirge_287 0.003290 FJ558066.1.1408 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;uncultured bacterium 97.5 1408 -1 97.5 1408 1408 0 35 1373
tongue.PFemirge_2259 0.003137 FJ470581.1.1490 Bacteria;Firmicutes;Clostridia;Clostridiales;Lachnospiraceae;Lachnospiraceae UCG-008;uncultured bacterium 99.4 1490 -1 99.4 1490 1490 0 9 1481
tongue.PFemirge_44 0.003105 AWSC01000005.3443.4969 Bacteria;Actinobacteria;Actinobacteria;Actinomycetales;Actinomycetaceae;Actinomyces;Actinomyces graevenitzii F0530 99.7 1519 -1 99.7 1519 1519 0 4 1515
tongue.PFemirge_447 0.003031 FJ983043.1.1537 Bacteria;Firmicutes;Clostridia;Clostridiales;Peptostreptococcaceae;Peptoclostridium;uncultured bacterium 90.3 1530 -1 90.9 1523 1516 14 134 1382
tongue.PFemirge_220 0.003019 FJ470573.1.1497 Bacteria;Firmicutes;Negativicutes;Selenomonadales;Veillonellaceae;Selenomonas;uncultured bacterium 99.1 1492 -1 99.0 1491 1490 2 12 1478
tongue.PFemirge_9891 0.002888 EU531782.1.1545 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;uncultured bacterium 78.6 1557 -1 82.2 1538 1485 72 261 1224
tongue.PFemirge_477 0.002708 HQ799343.1.1445 Bacteria;Firmicutes;Clostridia;Clostridiales;Lachnospiraceae;Lachnoanaerobaculum;uncultured organism 96.0 1438 -1 96.5 1431 1425 13 45 1380
tongue.PFemirge_381 0.002644 HE681253.1.1518 Bacteria;Firmicutes;Clostridia;Clostridiales;Ruminococcaceae;Ruminococcaceae UCG-014;uncultured bacterium 99.5 1508 -1 99.4 1508 1508 0 8 1500
tongue.PFemirge_195 0.002638 KF092280.1.1374 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;uncultured bacterium 93.9 1162 -1 94.8 1161 1158 4 67 1091
tongue.PFemirge_671 0.002544 AJ006963.1.1453 Bacteria;Firmicutes;Clostridia;Clostridiales;Family XIII;[Eubacterium] nodatum group;[Eubacterium] sulci 97.7 1453 -1 97.6 1491 1453 0 34 1419
tongue.PFemirge_1497 0.002483 JF189706.1.1349 Bacteria;Firmicutes;Clostridia;Clostridiales;Lachnospiraceae;Oribacterium;uncultured bacterium 97.6 1344 -1 97.5 1344 1344 0 32 1312
tongue.PFemirge_813 0.002332 KF101495.1.1358 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Porphyromonadaceae;Porphyromonas;uncultured bacterium 82.5 1331 -1 83.9 1323 1308 23 210 1098
tongue.PFemirge_779 0.002305 EF404014.1.1511 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;uncultured bacterium 96.6 1416 -1 97.3 1416 1406 10 38 1368
tongue.PFemirge_9890 0.002145 HQ778326.1.1469 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;uncultured organism 95.0 1467 -1 95.6 1461 1455 12 61 1394
tongue.PFemirge_1246 0.002121 HQ805205.1.1478 Bacteria;Firmicutes;Negativicutes;Selenomonadales;Veillonellaceae;Veillonella;uncultured organism 96.0 1479 -1 96.0 1478 1477 2 57 1420
tongue.PFemirge_339 0.002115 FJ557635.1.1415 Bacteria;Firmicutes;Negativicutes;Selenomonadales;Veillonellaceae;Megasphaera;uncultured bacterium 96.7 1415 -1 96.7 1415 1415 0 46 1369
tongue.PFemirge_147 0.002077 KF101349.1.1358 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella;uncultured bacterium 98.9 1358 -1 98.9 1358 1358 0 15 1343
tongue.PFemirge_247 0.001999 AF432138.1.1487 Bacteria;Fusobacteria;Fusobacteriia;Fusobacteriales;Leptotrichiaceae;Leptotrichia;Leptotrichia sp. oral clone FP036 99.1 1485 -1 99.1 1485 1485 0 13 1472
tongue.PFemirge_1087 0.001934 JF189684.1.1343 Bacteria;Actinobacteria;Actinobacteria;Micrococcales;Micrococcaceae;Rothia;uncultured bacterium 96.8 1347 -1 97.1 1343 1339 8 35 1304
tongue.PFemirge_9889 0.001919 AEVD01000030.72.1605 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;Streptococcus infantis ATCC 700779 84.6 1557 -1 88.4 1538 1484 73 167 1317
tongue.PFemirge_95 0.001868 FJ557743.1.1389 Bacteria;Firmicutes;Clostridia;Clostridiales;Lachnospiraceae;Stomatobaculum;uncultured bacterium 97.2 1368 -1 97.2 1368 1368 0 38 1330
tongue.PFemirge_235 0.001708 FJ470591.1.1499 Bacteria;Actinobacteria;Actinobacteria;Actinomycetales;Actinomycetaceae;Actinomyces;uncultured bacterium 98
PhyloFlash is going to try to reconstruct the taxonomic RNA subunits using three different methods. First by mapping the reads against a curated 16S rRNA SILVA database, then using the mapped reads it will try to reconstruct the sequences using SPAdes and EMIRGE.
From the previous file we can see that phyloFlash predicts around 200 OTUs in total. It also try to sort out the different taxa per abundance, based on the concept that the more a taxa is present in a sample the more sequenced reads it will be.
We know now that we should expect large diversity. From the different prediction methods it seems that even if the precise taxonomic identification varies, Betaproteobacteria, Bacterioidetes, Firmicutes and Gammaproteobacteria are among the most abundant taxa.
On the right is a pie chart representing the class of different OTU predicted by the EMIRGE module. I only displayed the taxa predicted with a ratio above 0,5% of the total reads used. This is certainly biased but it help predicting what is in our library.
Once this step is done, we can proceed to the next phase, the initial assembly and its analysis. This will be explained in the next article.